Software and Publications
We broadcast research in GitHub, scientific journals and web servers.
Software tools
We developed a number of software tools for genomic data analysis. Check them out at WGLab GitHub.
Full publication list
- The Google Citation report for the PI can be accessed here (Total citation: >55,000; H-index: 80). In 2022, the citation for the ANNOVAR (2010) paper reached 10,000, and InterVar (2017) reached 600.
Pre-prints
- Wu D, Yang J, Wang K. Multimodal Machine Learning Combining Facial Images and Clinical Texts Improves Diagnosis of Rare Genetic Diseases. arXiv, arXiv:2312.15320 [q-bio.QM]
- Wu D, Yang J, Wang K. Not All Large Language Models (LLMs) Succumb to the "Reversal Curse": A Comparative Study of Deductive Logical Reasoning in BERT and GPT Models. arXiv, arXiv:2312.03633 [cs.CL]
- Fang L, Chen Q, Wei CH, Lu Z, Wang K. Bioformer: an efficient transformer language model for biomedical text mining. arXiv, arXiv:2302.01588 [cs.CL]
Selected Publications
- Lai W, Zhao Y, Chen Y, et al. Autism patient-derived SHANK2B Y29X mutation affects the development of ALDH1A1 negative dopamine neuron. Molecular Psychiatry, in press
- Gracia-Diaz C, Perdomo JE, Khan ME, Roule T, Disanza BL, Cajka GG, Lei S, Gagne AL, Maguire JA, Shalem O, Bhoj EJ, Ahrens-Nicklas RC, French DL, Goldberg EM, Wang K, Glessner JT, Akizu N. KOLF2.1J iPSCs carry CNVs associated with neurodevelopmental disorders. Cell Stem Cell, 31(3):288-289, 2024
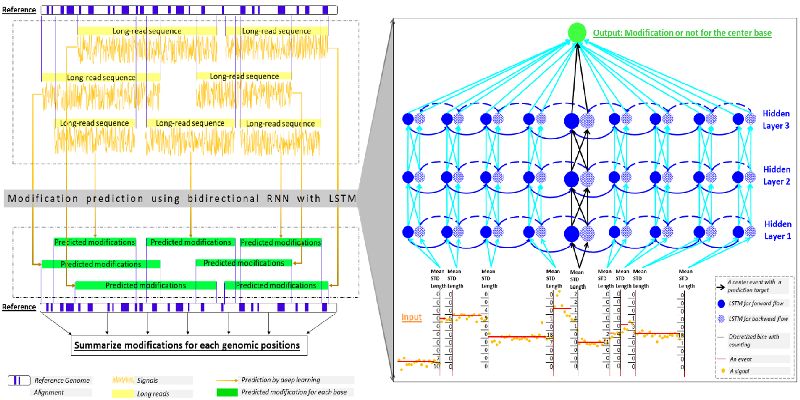
- Ahsan MU, Gouru A, Chan J, Zhou W, Wang K. A signal processing and deep learning framework for methylation detection using Oxford Nanopore sequencing. Nature Communications, 15(1):1448, 2024
- Gargano MA, Matentzoglu N, Coleman B, Addo-Lartey EB, Anagnostopoulos AV, et al. The Human Phenotype Ontology in 2024: phenotypes around the world. Nucleic Acids Research, 52(D1):D1333-D1346, 2024
- Murali H, Wang P, Liao EC, Wang K. Genetic variant classification by predicted protein structure: A case study on IRF6. Computational and Structural Biotechnology, 23:892-904, 2024
- Rybacki K, Xia M, Ahsan MU, Xing J*, Wang K*. Assessing the Expression of Long INterspersed Elements (LINEs) via Long-Read Sequencing in Diverse Human Tissues and Cell Lines. Genes, 14(10):1893, 2023
- Xu Z, Li Q, Marchionni L, Wang K. PhenoSV: interpretable phenotype-aware model for the prioritization of genes affected by structural variants. Nature Communications, 14(1):7805, 2023
- Yang J, Liu C, Deng W, Wu D, Weng C, Zhou Y, Wang K. Enhancing phenotype recognition in clinical notes using large language models: PhenoBCBERT and PhenoGPT. Patterns, 5(1):100887, 2023
- Jiang T, Fang L, Wang K. Deciphering the Language of Nature: A transformer-based language model for deleterious mutations in proteins. Innovation, 4:100487, 2023
- Wu D, Yang J, Ahsan MU, Wang K. Classification of integers based on residue classes via modern deep learning algorithms. Patterns, 4(12):100860, 2023
- Ahsan MU, Liu Q, Perdomo JE, Fang L, Wang K. A survey of algorithms for the detection of genomic structural variants from long-read sequencing data. Nature Methods, 20:1143–1158, 2023
- Wang X, Ahsan MU, Zhou Y, Wang K. Transformer-based DNA methylation detection on ionic signals from Oxford Nanopore sequencing data. Quantitative Biology, 11(3):287-296, 2023
- Lyon GJ, Vedaie M, Besheim T, Park A, Marchi E, et al. Expanding the Phenotypic spectrum of Ogden syndrome (NAA10-related neurodevelopmental syndrome) and NAA15-related neurodevelopmental syndrome. European Journal of Human Genetics, 31:824–833, 2023
- Fang L#, Mas Monteys A#, Dürr A, Keiser M, Cheng C, Harapanahalli A, Gonzalez-Alegre P, Davidson BL*, Wang K*. Haplotyping SNPs for allele-specific gene editing of the expanded huntingtin allele using long-read sequencing. HGG Advances, 4:100146, 2023
- Ren Z, Li Q, Cao K, Li MM, Zhou Y, Wang K. Model performance and interpretability of semi-supervised generative adversarial networks to predict oncogenic variants with unlabeled data. BMC Bioinformatics, 2023
- Li MM, Cottrell CE, Pullambhatla M, Roy S, Temple-Smolkin RL, Turner SA, Wang K, Zhou Y, Vnencak-Jones CL. Assessments of Somatic Variant Classification Using the Association for Molecular Pathology/American Society of Clinical Oncology/College of American Pathologists Guidelines: A Report from the Association for Molecular Pathology. Journal of Molecular Diagnostics, doi: 10.1016/j.jmoldx.2022.11.002, 2022
- Scott SA, Wang K, Spinner NB. Human Mutation special issue on innovations in genomic diagnostics. Human Mutation, 43(11):1493-1494, 2022
- Nixon A, Fang L, Havrilla JM, Wang K. Termviewer - A Web Application for Streamlined Human Phenotype Ontology (HPO) Tagging and Document Annotation. Chemistry and Biodiversity, 19:e202200805, 2022
- Li C, Zhi D, Wang K, Liu X. MetaRNN: Differentiating Rare Pathogenic and Rare Benign Missense SNVs and InDels Using Deep Learning. Genome Medicine, 14:115, 2022
- Chen Q, Allot A, Leaman R, Doğan RI, Du J, et al. Multi-label classification for biomedical literature: an overview of the BioCreative VII LitCovid Track for COVID-19 literature topic annotations. Database, 2022:baac069, 2022
- Doostparast Torshizi A, Wang K. Tissue-wide cell-specific proteogenomic modeling reveals novel candidate risk genes in autism spectrum disorders. npj Systems Biology and Applications, 8:31, 2022
- Liu C, Ta CN, Havrilla JM, Nestor JG, Spotnitz ME, Geneslaw AS, Hu Y, Chung WK, Wang K, Weng C. OARD: Open annotations for rare diseases and their phenotypes based on real-world data. American Journal of Human Genetics, 109:1591-1604, 2022
- Guo L, Park J, Yi E, Marchi E, Hsieh TC, Kibalnyk Y, Moreno-Sáez Y, Biskup S, Puk O, Beger C, Li Q, Wang K, Voronova A, Krawitz PM, Lyon GJ. KBG syndrome: videoconferencing and use of artificial intelligence driven facial phenotyping in 25 new patients. European Journal of Human Genetics, 30:1244–1254, 2022
- Havrilla JM, Singaravelu A, Driscoll DM, Minkovsky L, Helbig I, Medne L, Wang K, Krantz I, Desai BR. PheNominal: an EHR-integrated web application for structured deep phenotyping at the point of care, BMC Med Inform Decis Mak, 22:198, 2022
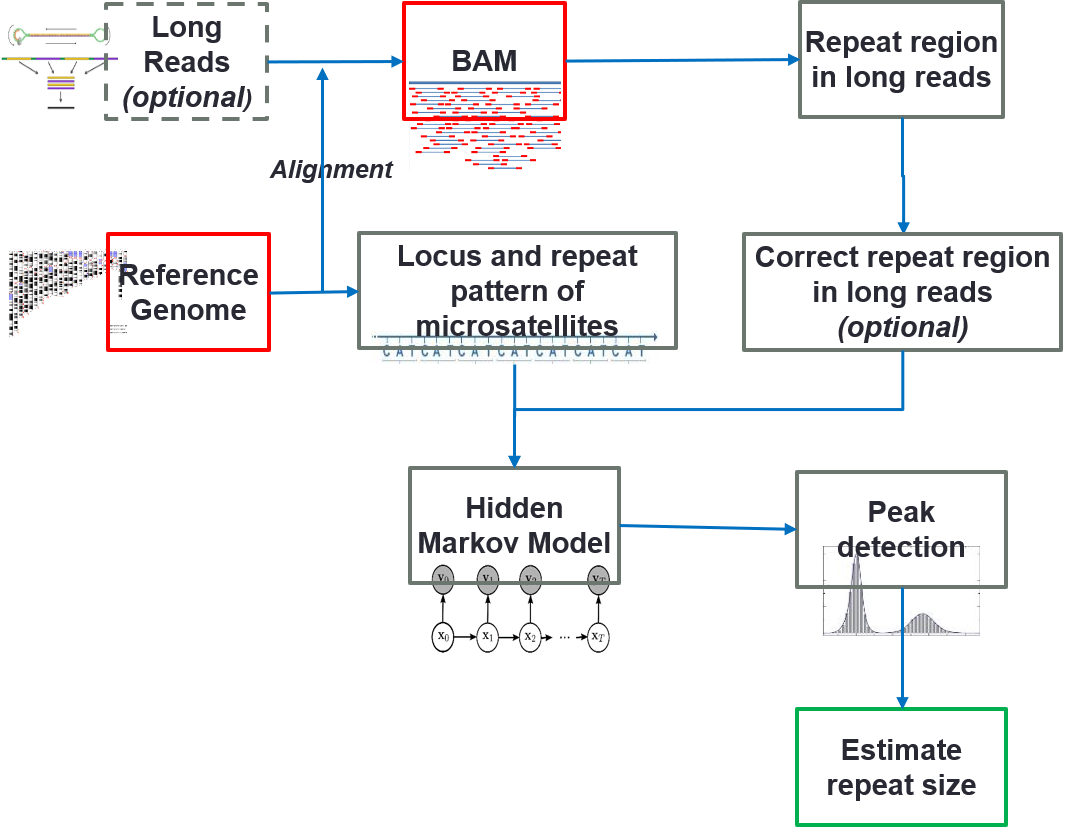
- Fang L#, Liu Q#, Monteys AM, Gonzalez-Alegre P, Davidson BL, Wang K. DeepRepeat: direct quantification of short tandem repeats on signal data from nanopore sequencing. Genome Biology, 23:128, 2022
- Fang L, Wang K. Polishing high-quality genome assemblies. Nature Methods, doi:10.1038/s41592-022-01515-1, 2022
- Olson ND, Wagner J, McDaniel J, et al. PrecisionFDA Truth Challenge V2: Calling variants from short and long reads in difficult-to-map regions. Cell Genomics, 2:100129, 2022.
- Zhao M, Havrilla J, Peng J, Drye M, Fecher M, Whitney Guthrie W, Tunc B, Schultz R, Wang K, Zhou Y. Development of a phenotype ontology for autism spectrum disorder by natural language processing on electronic health records. Journal of Neurodevelopmental Disorders, 14:32, 2022
- Wedemeyer MA, Muskens I, Strickland BA, Aurelio O, Martirosian V, Wiemels JL, Weisenberger DJ, Wang K, Mukerjee D, Rhie SK, Zada G. Epigenetic dysregulation in meningiomas. Neuro-Oncology Advances, 4:vdac084, 2022
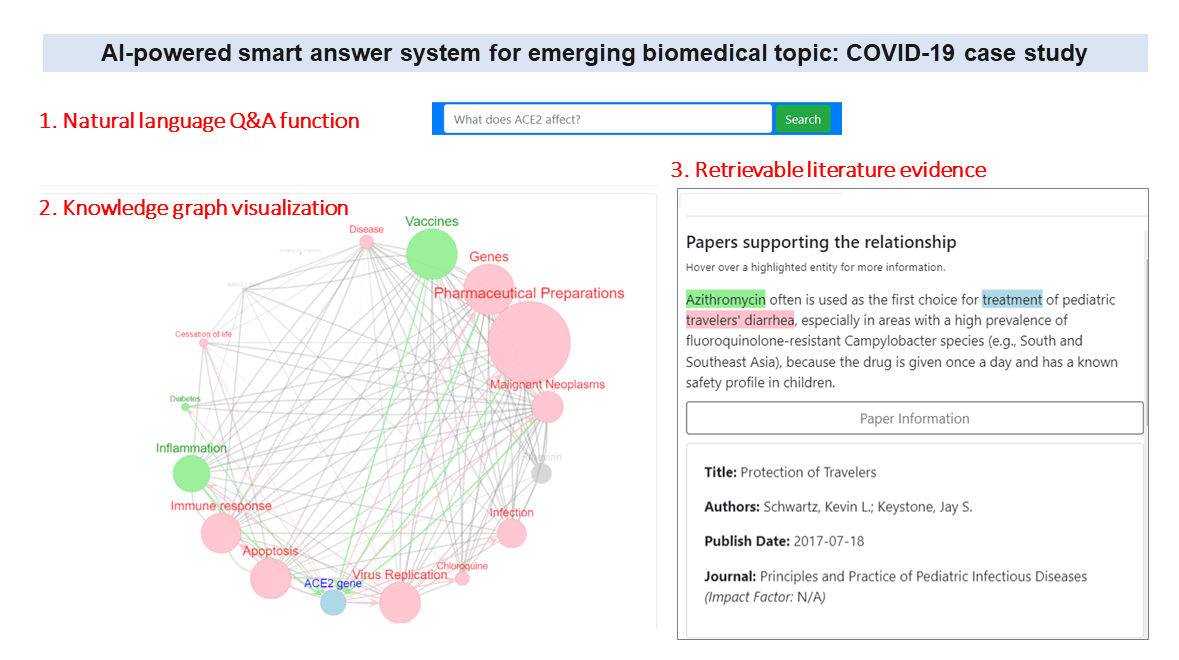
- Peng J, Xu D, Lee R, Xu S, Zhou Y, Wang K. Expediting knowledge acquisition by a web framework for Knowledge Graph Exploration and Visualization (KGEV): case studies on COVID-19 and Human Phenotype Ontology. BMC Medical Informatics and Decision Making, 22:147, 2022
- Li Q, Ren Z, Cao K, Li MM, Wang K*, Zhou Y*. CancerVar: An artificial intelligence–empowered platform for clinical interpretation of somatic mutations in cancer. Science Advances, 8(18), 2022.
- Ahsan U, Liu Q, Fang L, Wang K. NanoCaller for accurate detection of SNPs and small indels from long-read sequencing by deep neural networks. Genome Biology, 22(1):261, 2021.
- Havrilla J, Zhao M, Liu C, Weng C, Helbig I, Bhoj E, Wang K. Clinical Phenotypic Spectrum of 4095 Individuals with Down Syndrome from Text Mining of Electronic Health Records. Genes, 12(8):1159, 2021.
- Hu Y, Fang L, Chen X, Zhong JF, Li M, Wang K. LIQA: Long-read Isoform Quantification and Analysis. Genome Biology, 22: 182, 2021.
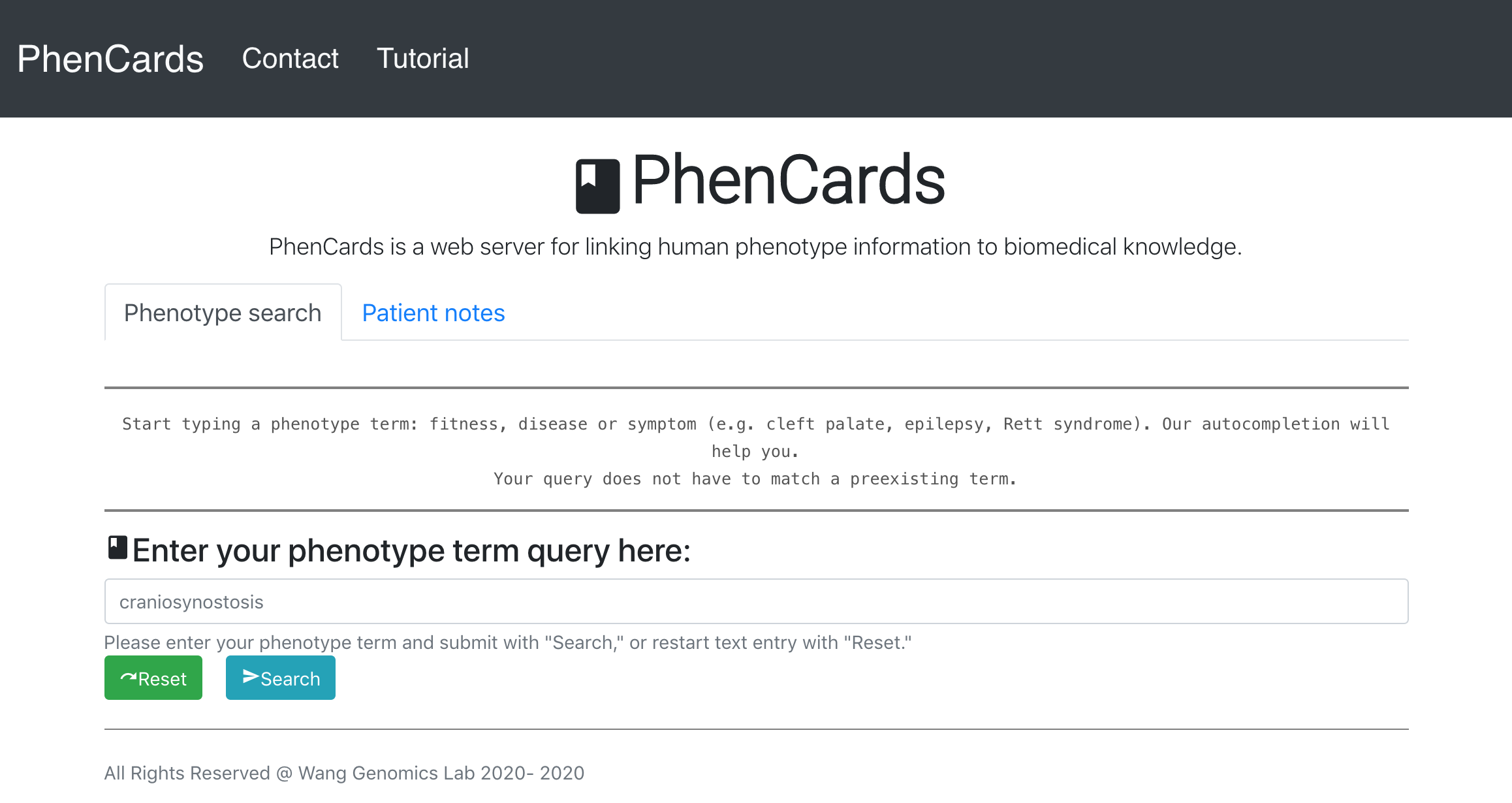
- Havrilla J, Liu C, Dong X, Weng C, Wang K. PhenCards: a data resource linking human phenotype information to biomedical knowledge. Genome Medicine, 13(1):91, 2021.
- Doostparast Torshizi A, Duan J, Wang K. A computational tool for direct inference of cell-specific expression profiles and cellular composition from bulk-tissue RNA-seq in brain disorders. NAR Genomics and Bioinformatics, 3(2):lqab056, 2021.
- Chen C, Yu W, Alikarami F, et al. Single-cell multiomics reveals increased plasticity, resistant populations and stem-cell-like blasts in KMT2A-rearranged leukemia. Blood, 2021.
- Ding X, Guo Y, Ye J, Wu X, Lin S, Chen F, Zhu L, Huang L, Song X, Zhang Y, Dai L, Xi X, Huang J, Wang K, Fan B, Li DW. Population differentiation and epidemic tracking of Bursaphelenchus xylophilus in China based on chromosome-level assembly and whole-genome sequencing data. Pest Management Science, doi:10.1002/ps.6738, 2021
- Huang H, Fang L, Liu Q, Doostparast Torshizi A, Wang K. Integrated analysis on transcriptome and behaviors defines HTT repeat-dependent network modules in Huntington's disease. Genes & Diseases, 2021.
- Doostparast Torshizi A, Duan J, Wang K. Cell type-specific proteogenomic signal diffusion for integrating multi-omics data predicts novel schizophrenia risk genes. Patterns, 1:100091, 2020.
- Liu Q, Hu Y, Stucky A, Fang L, Zhong JF, Wang K. LongGF: computational algorithm and software tool for fast and accurate detection of gene fusions by long-read transcriptome sequencing, BMC Genomics, 21:793, 2020.
- Hu Y, Fang L, Nicholson C, Wang K. Implications of error-prone long-read whole-genome shotgun sequencing on characterizing reference microbiomes. iScience, 23:101223, 2020.
- Doostparast Torshizi A, Ionita-Laza I, Wang K. Cell Type-Specific Annotation and Fine Mapping of Variants Associated With Brain Disorders. Frontiers in Genetics, 11: 575928, 2020
- Yang H, Luo Y, Liu T, et al. A map of cis-regulatory elements and 3D genome structures in zebrafish. Nature, 588:337–343, 2020.
- Huang D, Yi X, Zhou Y, Yao H, Xu H, Wang J, Zhang S, Nong W, Wang P, Shi L, Xuan C, Li M, Wang J, Li W, Kwan HS, Sham PC, Wang K, Li MJ. Ultrafast and scalable variant annotation and prioritization with big functional genomics data. Genome Research, 30:1789-1801, 2020.
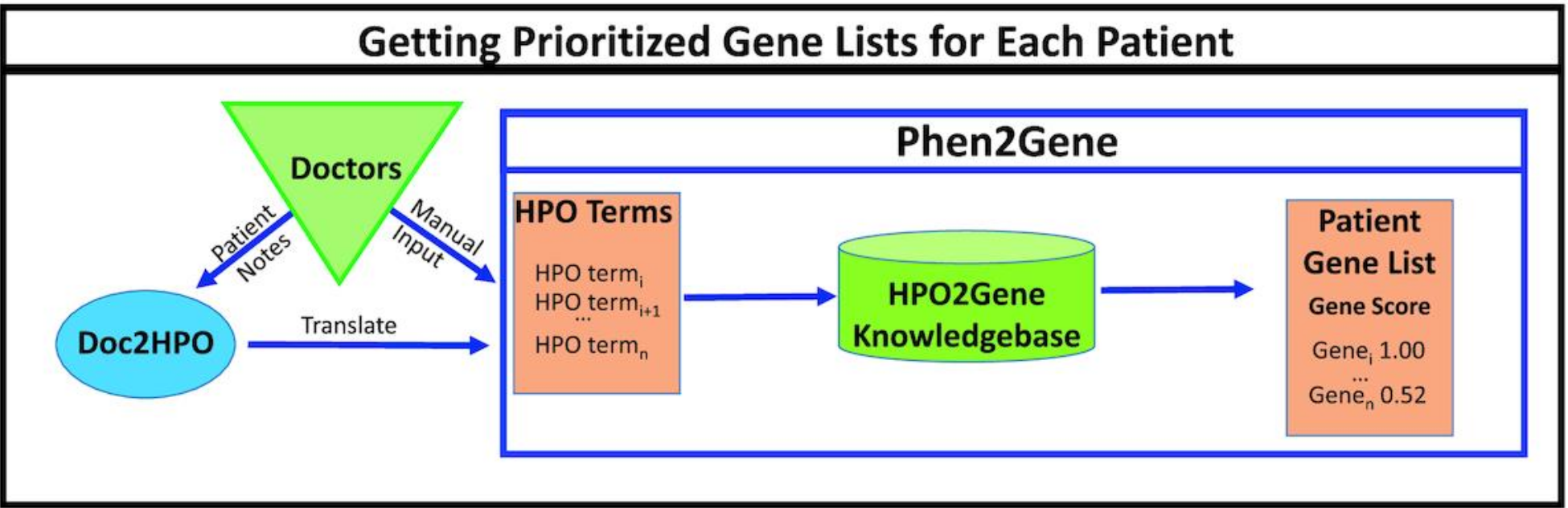
- Zhao M, Havrilla JM, Fang L, Chen Y, Peng J, Liu C, Wu C, Sarmady M, Botas P, Isla J, Lyon GJ, Weng C*, Wang K*. Phen2Gene: Rapid Phenotype-Driven Gene Prioritization for Rare Diseases. NAR Genomics and Bioinformatics, 2:lqaa032, 2020.
- Georgieva D, Liu Q, Wang K*, Egli D*. Detection of Base Analogs Incorporated During DNA Replication by Nanopore Sequencing. Nucleic Acids Research, 48:e88, 2020
- Hu Y, Wang K, Li M. Detecting differential alternative splicing events in scRNA-seq with or without Unique Molecular Identifiers. PLoS Computational Biology, 16:e1007925, 2020
- Liu Q, Tong Y, Wang K. Genome-wide detection of short tandem repeat expansions by long-read sequencing. BMC Bioinformatics, 21:542, 2020.
- Evgrafov OV, Armoskus C, Wrobel BB, Spitsyna VN, Souaiaia T, Herstein JS, Walker CP, Nguyen JD, Camarena A, Weitz JR, Kim JM, Duarte EL, Wang K, Simpson GM, Sobell JL, Medeiros H, Pato MT, Pato CN, Knowles JA: Gene Expression in Patient-Derived Neural Progenitors Implicates WNT5A Signaling in the Etiology of Schizophrenia. Biological Psychiatry, 88:236-247, 2020
- Wang L, Wang Q, Bai H, Liu C, Liu W, Zhang Y, Jiang L, Xu H, Wang K*, Zhou Y*. EHR2Vec: Representation Learning of Medical Concepts From Temporal Patterns of Clinical Notes Based on Self-Attention Mechanism, Frontiers in Genetics, 11:630, 2020.
- Peng J, Zhao M, Havrilla J, Liu C, Weng C, Guthrie W, Schultz R, Wang K*, Zhou Y*. Natural Language Processing (NLP) tools in extracting biomedical concepts from research articles: a case study on autism spectrum disorder. BMC Medical Informatics and Decision Making, 20:322, 2020
- Ling C, Dai Y, Fang Li, Yao F, Liu Z, Qiu Z, Cui L, Xia F, Zhao C, Zhang S, Wang K*, Zhang X*. Exonic rearrangements in DMD in Chinese Han individuals affected with Duchenne and Becker muscular dystrophies. Human Mutation, 41:668-677, 2020.
- Wu J, Li Y, Wang C, Cui Y, Xu T, Wang C, Wang X, Sha J, Jiang B, Wang K, Hu Z, Guo X, Song X. CircAST: Full-length Assembly and Quantification of Alternatively Spliced CircRNA Isoforms. Genomics Proteomics Bioinformatics, 17(5): 522-534, 2020
- Dai Y, Li P, Wang Z, Liang F, Yang F, Fang L, Huang Y, Huang S, Zhou J, Wang D, Cui L, Wang K: Single-molecule optical mapping enables quantitative measurement of D4Z4 repeats in facioscapulohumeral muscular dystrophy (FSHD). Journal of Medical Genetics, 57:109-120, 2020.
- Fang L, Kao C, Gonzalez MV, Mafra FA, Pellegrino da Silva R, Li M, Wenzel S, Wimmer K, Hakonarson H, Wang K. LinkedSV: Detection of mosaic structural variants from linked-read exome and genome sequencing data. Nature Communications, 10:5585, 2019.
- Doostparast Torshizi A, Armoskus C, Zhang H, Forrest MP, Zhang S, Souaiaia T, Evgrafov OV, Knowles JA, Duan J*, Wang K*: Deconvolution of Transcriptional Networks Identified TCF4 as a Master Regulator in Schizophrenia. Science Advances, 5:eaau4139, 2019.
- Liu C, Peres Kury FS, Li Z, Ta C, Wang K*, Weng C*. Doc2Hpo: a web application for efficient and accurate HPO concept curation. Nucleic Acids Research, 47:W566-W570, 2019
- He MM, Li Q, Yan M, Cao H, Hu Y, He KY, Cao K, Li MM, Wang K. Variant Interpretation for Cancer (VIC): a computational tool for assessing clinical impacts of somatic variants. Genome Medicine, 11:53, 2019
- Liu Q, Fang L, Yu G, Wang D, Xiao CL*, Wang K*. Detection of DNA base modifications by deep recurrent neural network on Oxford Nanopore sequencing data. Nature Communications, 10:2449, 2019
- Xie G, Dong C, Kong Y, Zhong JF, Li M, Wang K. GDP: Group lasso regularized Deep learning for cancer Prognosis from multi-omics and clinical features. Genes, 10:240, 2019.
- Khan A, Liu Q, Chen X, Zeng Y, Stucky A, Sedghizadeh PP, Adelpour D, Zhang X, Wang K*, Zhong JF*: Detection of human papillomavirus in cases of head and neck squamous cell carcinoma by RNA-seq and VirTect. Molecular Oncology, 13:829-839, 2019
- Zeng S, Zhang MY, Wang XJ, Hu ZM, Li JC, Li N, Wang JL, Liang F, Yang Q, Liu Q, Fang L, Hao JW, Shi FD, Ding XB, Teng JF, Yin XM, Jiang H, Liao WP, Liu JY, Wang K*, Xia K*, Tang BS*: Long-read sequencing identified intronic repeat expansions in SAMD12 from Chinese pedigrees affected with Familial Cortical Myoclonic Tremor with Epilepsy. Journal of Medical Genetics, 56:265-270, 2019
- Borgmann-Winter KE, Wang K, Bandyopadhyay S, Doostparast Torshizi A, Blair I, Hahn CY. The proteome and its dynamics: A missing piece for integrative multi-omics in schizophrenia. Schizophrenia Research, doi: 10.1016/j.schres.2019.07.025, 2019
- Paine I, Posey JE, Grochowski CM, Jhangiani SN, Rosenheck S, et al. Paralog Studies Augment Gene Discovery: DDX and DHX Genes. American Journal of Human Genetics, 105(2):302-316, 2019
- Lyon GJ, Marchi E, Ekstein J, Meiner V, Hirsch Y, Scher S, Yang E, De Vivo DC, Madrid R, Li Q, Wang K, Haworth A, Chilton I, Chung WK, Velinov M. VAC14 syndrome in two siblings with retinitis pigmentosa and neurodegeneration with brain iron accumulation. Cold Spring Harbor Molecular Case Studies, 5(6):a003715, 2019
- Liu Q, Shi L, Wang K. Ethnicity-Specific Reference Genome Assembly by Long-Read Sequencing. J Mol Genet Med, 12:385, 2018
- Liu Q, Georgieva DC, Egli DM, Wang K. NanoMod: a computational tool to detect DNA modifications using Nanopore long-read sequencing data. BMC Genomics, 20:78, 2018
- Chen Y, Millstein J, Liu Y, Chen GY, Chen X, Stucky A, Qu C, Fan JB, Chang X, Soleimany A, Wang K, Zhong J, Liu J, Gilliland FD, Li Z, Zhang X, Zhong JF. Single-Cell Digital Lysates Generated by Phase-Switch Microfluidic Device Reveal Transcriptome Perturbation of Cell Cycle. ACS Nano, 12:4687-4694, 2018
- Khan A, Liu Q, Wang K. iMEGES: integrated Mental-disorder GEnome score for prioritizing the susceptibility genes for mental disorders in personal genomes. BMC Bioinformatics, 19:501, 2018
- He Z, Liu L, Wang K, Ionita-Laza I. A semi-supervised approach for predicting cell type specific functional consequences of non-coding variation using MPRAs. Nature Communications, 9:5199, 2018
- Xiao CL, Zhu S, He M-H, Chen Y, Yu GL, Chen D, Xie SQ, Luo F, Liang Z, Wang DP, Bo XC*, Gu XF*, Wang K*, Yan GR*. N6-methyladenine DNA modification in human genome. Molecular Cell, 71:306-318, 2018
- Hoon Son J, Xie G, Yuan C, Ena L, Li Z, Goldstein A, Huang L, Wang L, Shen F, Liu H, Mehl K, Groopman EE, Marasa M, Kiryluk K, Gharavi AG, Chung WK, Hripcsak G, Friedman C, Weng C*, Wang K*. Deep phenotyping on electronic health records facilitates genetic diagnosis by clinical exomes. American Journal of Human Genetics, 103:58-73, 2018
- Doostparast Torshizi A, Wang K. Next-generation sequencing in drug development: target identification and genetically stratified clinical trials. Drug Discovery Today, 23:1776-1783, 2018
- Fang L, Hu J, Wang D, Wang K. NextSV: a computational pipeline for structural variation analysis from low-coverage long-read sequencing. BMC Bioinformatics, 19:180, 2018
- Miao H, Zhou J, Yang Q, Liang F, Wang D, Ma N, Gao B, Du J, Lin G, Wang K*, Zhang Q*. Long-read sequencing identified a causal structural variant in an exome-negative case and enabled preimplantation genetic diagnosis. Heraditas, 115:32, 2018
- Khan A, Wang K. A deep learning based scoring system for prioritizing susceptibility variants for mental disorders. IEEE International Conference on Bioinformatics and Biomedicine. Page: 1698-1705, DOI: 10.1109/BIBM.2017.8217916, 2017
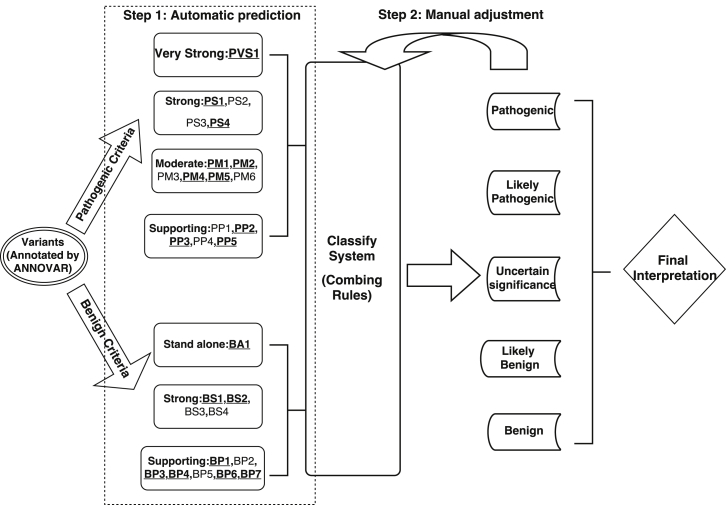
- Li Q, Wang K. InterVar: Clinical interpretation of genetic variants by ACMG/AMP 2015 guidelines, American Journal of Human Genetics, 100:267-280, 2017
- Liu Q, Zhang P, Wang D, Gu W, Wang K. Interrogating the "unsequenceable" genomic trinucleotide repeat disorders by long-read sequencing. Genome Medicine, 9:65, 2017
- Li J, Zhang W, Yang H, Howrigan DP, Wilkinson B, Souaiaia T, Evgrafov OV, Genovese G, Clementel VA, Tudor JC, Abel T, Knowles JA, Neale BM, Wang K, Sun F, Coba MP: Spatiotemporal profile of postsynaptic interactomes integrates components of complex brain disorders. Nature Neuroscience, 20:1150-1161, 2017
- de Araújo Lima LA, Wang K: PennCNV in whole-genome sequencing data. BMC Bioinformatics, 18:383, 2017
- Dong C, Guo Y, Yang H, He Z, Liu X, Wang K. iCAGES: integrated CAncer GEnome Score for comprehensively prioritizing driver genes in personal cancer genomes, Genome Medicine, 8:135, 2016
- Shi L, Guo Y, Dong C, Huddleston J, Yang H, Han X, Fu A, Li Q, Li N, Gong S, Lintner KE, Ding Q, Wang Z, Hu J, Wang D, Wang F, Wang L, Lyon GJ, Guan Y, Shen Y, Evgrafov OV, Knowles JA, Thibaud-Nissen F, Schneider V, Yu CY, Zhou L, Eichler EE, So KF, Wang K. Long read sequencing and de novo assembly of a Chinese genome. Nature Communications, 7:12065, 2016
- Cai M, Gao F, Lu W, Wang K. w4CSeq: software and web application to analyze 4C-Seq data, Bioinformatics, 32:3333-3335, 2016
- Song X, Zhang N, Han P, Lai RK*, Wang K*, Lu W*. Circular RNA Profile in Gliomas Revealed by Identification Tool UROBORUS. Nucleic Acids Research, 44:e87, 2016
- He KY, Zhao Y, McPherson EW, Li Q, Xia F, Weng C, Wang K*, He MM*. Pathogenic Mutations in Cancer-Predisposing Genes: A Survey of 300 Patients with Whole-Genome Sequencing and Lifetime Electronic Health Records. PLoS One, 11:e0167847, 2016
- Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, et al. The PsychENCODE Project, Nature Neuroscience, 18:1707-1712, 2015
- Guo Y, Ding X, Shen Y, Lyon GJ, Wang K. SeqMule: automated analysis pipeline for analysis of human exome/genome sequencing data. Scientific Reports, 5:14283, 2015
- Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nature Protocols, 10:1556-1566, 2015
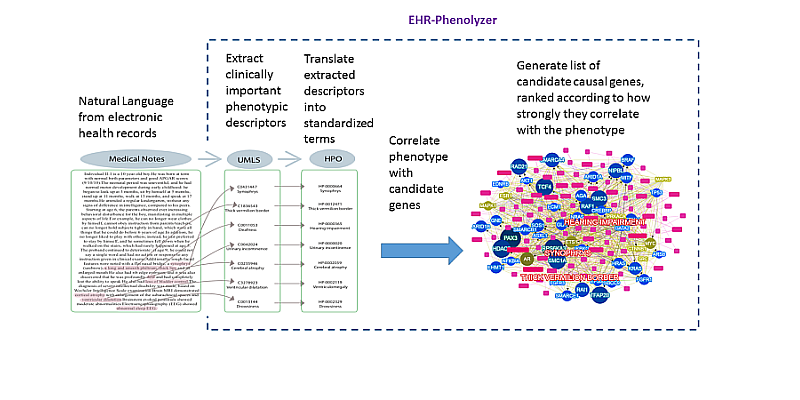
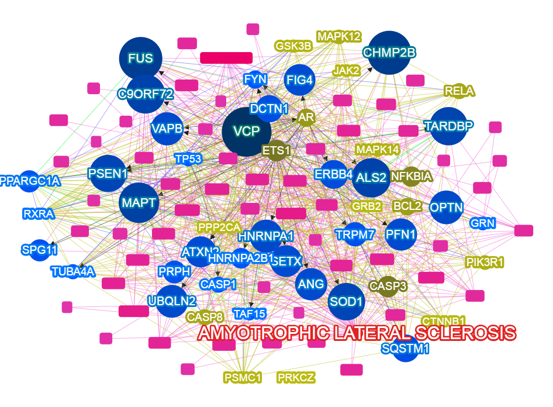
- Yang H., Robinson PN, Wang K. Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nature Methods, 12:841-843, 2015
- He M, Person TN, Hebbring SJ, Heinzen E, Ye Z, Schrodi SJ, McPherson EW, Lin SM, Peissig PL, Brilliant MH, O'Rawe J, Robison RJ, Lyon GJ, Wang K. SeqHBase: a big data toolset for family-based sequencing data analysis. Journal of Medical Genetics, 52:282-288, 2015
- Dong C, Wei P, Jian X, Gibbs R, Boerwinkle E, Wang K*, Liu X*. Comparison and integration of deleteriousness prediction methods of nonsynonymous SNPs in whole exome sequencing studies. Human Molecular Genetics, 24:2125-2137, 2015
- Guo Y, Conti DV, Wang K. Enlight: web-based integration of GWAS results with biological annotations. Bioinformatics, 31:275-276, 2015
- Gao F, Wang K. Ligation-anchored PCR unveils immune repertoire of TCR-beta from whole blood. BMC Biotechnology, 15:39, 2015
- Jia H, Guo Y, Zhao W, Wang K. Long-range PCR in next-generation sequencing: comparison of six enzymes and evaluation on the MiSeq sequencer. Scientific Reports, 4:5737, 2014
- Shi L, Li B, Huang YL, Ling XY, Liu T, Lyon GJ, Xu A, Wang K. "Genotype-first" approaches on a curious case of idiopathic progressive cognitive decline. BMC Medical Genomics, 7:66, 2014
- Gao F, Wei Z, Lu W, Wang K. Comparative analysis of 4C-Seq data generated from enzyme-based and sonication-based methods. BMC Genomics, 14:345, 2013
- Wei Z, Gao F, Kim S, Yang H, Wang K, Lu W. Klf4 Organizes Long-Range Chromosomal Interactions with the Oct4 Locus in Reprogramming and Pluripotency. Cell Stem Cell, 13:36-47, 2013
- Chen G, Chang X, Curtis C, Wang K. Precise inference of copy number alterations from SNP arrays. Bioinformatics, 29:2964-2970, 2013
- Wang K*, Kim C, Bradfield J, Guo Y, Toskala E, Otieno FG, Hou C, Thomas K, Cardinale C, Lyon GL, Golhar R, Hakonarson H*. Whole-genome DNA/RNA sequencing on a novel Mendelian disease with neuromuscular and cardiac involvement. Genome Medicine, 5:67, 2013
- Shi L, Chang X, Zhang P, Coba M, Lu W, Wang K. The functional genetic link of NLGN4X knockdown and neurodevelopment in neural stem cells. Human Molecular Genetics, 22:3749:3760, 2013
- Gao F, Ling C, Shi L, Commins D, Zada G, Mack W, Wang K. Inversion-mediated gene fusion involving NAB2-STAT6 in an unusual malignant meningioma. British Journal of Cancer, 109:1051-1055, 2013
- Gao F, Shi L, Russin J, Zeng L, Chang X, He S, Chen TC, Giannotta SL, Weisenberger DJ, Zada G, Mack WJ, Wang K. DNA methylation in the malignant transformation of meningiomas. PLoS ONE, 8:e54114, 2013
- Chang X, Xu T, Li Y, Wang K. Dynamic modular architecture of protein-protein interaction networks beyond the dichotomy of 'date' and 'party' hubs. Scientific Reports, 3:1691, 2013
- Qiu S, Luo S, Evgrafov O, Li R, Schroth GP, Levitt P, Knowles JA*, Wang K*. Single-neuron RNA-Seq: technical feasibility and reproducibility. Frontiers in Genetics, 3:124, 2012
- Lyon GJ, Wang K. Identifying disease mutations in genomic medicine settings: current challenges and how to accelerate progress. Genome Medicine, 4:58, 2012
- Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. Journal of Medical Genetics. 49:433-436, 2012
- Lyon GJ, Jiang T, Van Wijk R, Wang W, Bodily P, Xing J, Tian L, Robison R, Clement M, Yang L, Zhang P, Liu Y, Moore B, Glessner J, Elia J, Reimherr F, van Solinge W, Yandell M, Hakonarson H, Wang J, Johnson WE, Wei Z, Wang K. Exome Sequencing and Unrelated Findings in the context of Complex Disease Research: Ethical and Clinical Implications. Discovery Medicine, 12:41-55, 2011
- Wang K*, Diskin SJ*, Zhang H*, Attiyeh EF, Winter C, Hou C, Schnepp RW, Diamond M, Bosse K, Mayes PA, Glessner J, Kim C, Frackelton E, Garris M, Wang Q, Glaberson W, Chiavacci R, Nguyen L, Jagannathan J, Saeki N, Sasaki H, Grant SF, Iolascon A, Mosse YP, Cole KA, Li H, Devoto M, McGrady PW, London WB, Capasso M, Rahman N, Hakonarson H, Maris JM. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature, 469:216-220, 2011
- Wang K, Zhang H, Bloss CT, Duvvuri V, Kaye W, Schork NJ, Berrettini W, Hakonarson H, the Price Foundation Collaborative Group. A genome-wide association study on common SNPs and rare CNVs in anorexia nervosa. Molecular Psychiatry, 16:949-959, 2011
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research, 38:e164 2010
- Wang K, Li M, Hakonarson H. Analysing biological pathways from genome-wide association studies. Nature Reviews Genetics, 11:843-854, 2010
- Wang K, Bucan M, Grant SF, Schellenberg G, Hakonarson H. Strategies for genetic studies of complex diseases. Cell, 142:351-353, 2010
- Wang K, Baldassano R, Zhang H, et al. Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effects. Human Molecular Genetics, 19:2059-2967, 2010
- Wang K*, Zhang H*, Ma D*, Bucan M, Glessner JT, Abrahams BS, Salyakina D, Imielinski M, Bradfield JP, Sleiman PM, Kim CE, Hou C, Frackelton E, Chiavacci R, Takahashi N, Sakurai T, Rappaport E, Lajonchere CM, Munson J, Estes A, Korvatska O, Piven J, Sonnenblick LI, Alvarez Retuerto AI, Herman EI, Dong H, Hutman T, Sigman M, Ozonoff S, Klin A, Owley T, Sweeney JA, Brune CW, Cantor RM, Bernier R, Gilbert JR, Cuccaro ML, McMahon WM, Miller J, State MW, Wassink TH, Coon H, Levy SE, Schultz RT, Nurnberger JI, Haines JL, Sutcliffe JS, Cook EH, Minshew NJ, Buxbaum JD, Dawson G, Grant SF, Geschwind DH, Pericak-Vance MA, Schellenberg GD, Hakonarson H. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature, 459:528-533, 2009
- Wang K, Horst JA, Cheng G, Nickle DC, Samudrala R. Protein meta-functional signatures from combining sequence, structure, evolution, and amino acid property information. PLoS Computational Biology, 4:e1000181, 2008
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant S, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Research, 17:1665-1674, 2007
- Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genome-wide association studies. American Journal of Human Genetics, 81:1278-1283, 2007
- Wang K, Mittler JE, Samudrala R. Comment on Evidence for positive epistasis in HIV-1. Science, 312:848, 2006
- Wang K. Gene-function wiki would let biologists pool worldwide resources. Nature, 439:534, 2006
- Wang K, Samudrala R. Incorporating background frequency improves entropy-based residue conservation measures. BMC Bioinformatics, 17:385, 2006
- Wang K, Samudrala R. FSSA: a novel method for identifying functional signatures from structural alignments. Bioinformatics, 21:2969-2977, 2005
Book Chapters
- Guan Y, Wang K. Whole-genome multi-SNP analysis. In: Statistical Bioinformatics. Edited by Do KA, Qin Z, Vannucci M. Cambridge University Press, 2013
- Wang K. Epistasis. In: Encyclopedia of Autism Spectrum Disorders. Edited by Volkmar FR. Springer, 2013
- Fang L, Wang K. Identification of Copy Number Variants from SNP Arrays Using PennCNV. In: Methods in Molecular Biology. Edited by Derek Bickhart. Springer, vol. 1833, 2018
Software/Web server